Supervised ML - Regression (I)
Machine Learning is a difficult concept, but a necessary one that will open you to a new world of possibilities in data analysis. To get up to speed quickly, you will need to learn theory, terminology, core algorithms, and application.
Here’s the roadmap for machine learning regression:
- Linear Models
- Linear Regression
- Generalized Linear Models (GLM) - Elastic Net
- Tree-Based Models
- Decision Trees
- Random Forest
- XGBoost (Gradient Boosted Machine, GBM)
- Support Vector Machine & Preprocessing
With this roadmap, you will be able to learn the basics of modeling using all of the core algorithms. Further, you will understand how they work along with their strengths and weaknesses.
ML Theory
Machine Learning: A Gentle Introduction
This is an excellent tutorial covering basic concepts of machine learning.
Bias versus Variance: Fighting Over-Fitting
The Bias/Variance tradeoff is why we need to test the prediction accuracy on unseen data. This tutorial forms our intuition for separating our data into training and testing data sets.
Cross Validation
Summary
- Common Applications in Business: Used to predict a numeric value (e.g. forecasting sales, estimating prices, etc).
- Key Concept: Data is usually in a rectangular format (like a spreadsheet) with one column that is a target (e.g. price) and other columns that are predictors (e.g. product category)
- Gotchas:
- Preprocessing: Knowing when to preprocess data (normalize) prior to machine learning step
- Feature Engineering: Getting good features is more important than applying complex models.
- Parameter Tuning: Higher complexity models have many parameters that can be tuned.
- Interpretability: Some models are more explainable than others, meaning the estimates for each feature means something in relation to the target. Other models are not interpretable and require additional tools (e.g. LIME) to explain.
Terminology
- Supervised vs Unsupervised: Regression is a supervised technique that requires training with a “target” (e.g. price of product or sales by month). The algorithm learns by identifying relationships between the target & the predictors (attributes related to the target like category of product or month of sales).
- Classification vs Regression: Classification aims to predict classes (either binary yes/no or multi-class categorical). Regression aims to predict a numeric value (e.g. product price = $4,233).
- Preprocessing: Many algorithms require preprocessing, which transforms the data into a format more suitable for the machine learning algorithm. A common example is “standardization” or scaling the feature to be in a range of [0,1] (or close to it).
- Hyper Parameter & Tuning: Machine learning algorithms have many parameters that can be adjusted (e.g. learning rate in GBM). Tuning is the process of systematically finding the optimum parameter values.
- Cross Validation: Machine learning algorithms should be tuned on a validation set as opposed to a test set. Cross-validation is the process of splitting the training set into multiple sets using a portion of the training set for tuning.
- Performance Metrics (Regression): Common performance metrics are Mean Absolute Error (MAE) and Root Mean Squared Error (RMSE). These measures provide an estimate of model performance to compare models to each other.
Machine Learning Algorithms - Regression
| Popular Algorithms | Type | Key Concepts | Feature Range Standardization [0,1] | Results interpretable? | Key Parameters |
|---|---|---|---|---|---|
| Linear Regression | Linear | Simplest method - Uses OLS to reduce error | Not required | Yes - Model terms indicate magnitude / direction of each feature contribution | N/A |
| GLM (Generalized Linear Model)LASSO, Rdige Regression, Elastic Net | Linear | Linear method that penalizes irrelevant features using a concept callend “Regularization”, where the weight of the irrelevant featues is reduced to make their effect on the model lower.L1 Regularization - Called LASSO regressionL2 regularization - Called Ridge RegressionElastic Net - Combines L1 & L2 Regularization | Required.1 | Yes, if standardization is performed internally to algorithm. Model terms indicate magnitude / direction of each features contribution | Penalty (alpha) - How much to penalize the paramentersMixture (L1 Ratio) - Ratio between L1 and L2 regularization |
| Decision Tree | Tree-Based (Non-Linear) | A decision tree is a set of decision rules. Each rule is considered a node with a split being a binary decision. The decisions terminate at a leaf. | Not required | Yes - Decision Tree Plots show rule-based decisions that show how to arrive at model prediction | Max Tree Depth - How many splits for the longest treeMin Samples per leaf / Node - How many samples on each end node (leaf)Cost Complexity (Cp) / Min impurity - Instructs when to stop (create a leaf) if additional information gain is not above a Cp threshold |
| Random Forest | Tree-Based (Non-Linear) | Ensemble learning method where many trees are created on sub-samples of data set and combined using averaging. This process controls overfitting, typicallying leading to a more accurate model. However, because the models are combined, the ecision rules become incomprehensible. This process is often called “Bagging”. | Not required | No2 | See Decision Trees Parameters, and:Replacement - whether or not to draw samples with replacementNumber of features - How many columns to use when samplingNumber of trees - How many trees to average |
| GBM (Gradient Boosted Machine)XGBoost | Tree-Based (Non-Linear) | Implements a technique called “Boosting” to build decision trees of weak prediction models and generalizes using a loss function. The weak learners converge to a strong learner. | Not required | No2 | See RandomForest Key Parameters, and:Learning Rate (eta) - The rate that the boosting algorithms adaptsLoss Reduction (gamma) - The loss function to use during splittingSample Size - The proportion of data exposed to the model during each iteration |
| SVM (Support Vector Machines) | Kernel Basis (Polynomial or Radial) (Non-Linear) | An algorithm that uses a kernel to transform the feature space to linearly seperable boundaries, and then applies a margin penalizing points that are incorrectly measured outside of the margin. The kernel transformation (i.e. radial, polynomial) makes it possible to perform linear separations within non-linear data. | Required3 | No2 | Kernel - Ploynomial or Radial Basis functionCost / Regularization - Cost of predicting sample on wrong side of the SVM marginMargin (epsilon) - Specifies region where no penalty is appliedDegree (Polynomial) - Degree of Polynomial. Use 1 for linear, 2 or more for flexible (quadratic)Scale factor (Polynomial) - Factor to adjust bias/varianceGamma or Sigma (Radial) - Factor to adjust bias/variance |
| Deep Learning (Neural Network) | Neural Network (Non-Linear) | Learning algorithms with input and output and layers in between where the model parameters are learned. The user develops the architecture of the neural network, and the algorithm learns the model through iteratively seeking to minimize a cost function. | Required | No2 | Many tuning parameters & architecture decisions |
Theory Input
The tidymodels framework is a collection of packages for modeling and machine learning using tidyverse principles. tidymodels offers a consistent, flexible framework for your work. Install many of the packages in the tidymodels ecosystem by running install.packages("tidymodels"). The core tidymodels packages work together to enable a wide variety of modeling approaches. Parsnip is the cornerstone modeling package, which is why I teach it (instead of many other packages that offer also a wide range of models and tools for modeling, e.g. caret). It is an interface that standardizes the making of models in R. You can connect many engines to parsnip. An engine is the underlying algorithm that your are connecting parsnip to (e.g. lm, glmnet or keras) . This will make more sense once we get to our modeling section.
rsampleprovides infrastructure for efficient data splitting, resampling and cross validation (we will use for randomly splitting data).parsnipprovides an API to many powerful modeling algorithms in R.recipesis a tidy interface to data pre-processing (making statistical transformations) tools for feature engineering (prior to modeling).workflowsbundle your pre-processing, modeling, and post-processing together.
tunehelps you optimize the hyperparameters of your model and pre-processing steps.yardstickmeasures the effectiveness of models using performance metrics (metrics for model comparison).broomconverts the information in common statistical R objects into user-friendly, predictable formats.dialscreates and manages tuning parameters and parameter grids.
Let’s learn what you need to get started with tidymodels in four steps, starting with how to create a model and ending with how to tune model parameters. The following examples are from that site.
1. Modeling
How do you create a statistical model using tidymodels? We start with data for modeling, learn how to specify and train models with different engines using the parsnip package, and understand why these functions are designed this way.
You will need the following packages:
library(tidymodels) # for the parsnip package, along with the rest of tidymodels
# Helper packages
library(broom.mixed) # for converting bayesian models to tidy tibbles
I. The data
I did some further data cleaning and added the weights and frame material for each model. See code below.
bike_data_sizes_tbl <- readRDS("~/bike_data_sizes_tbl.rds")
bike_data_tbl <- bike_data_sizes_tbl %>%
group_by_at(.vars = vars(name:description, url_base)) %>%
summarise(across(stock_availability, sum)) %>%
ungroup() %>%
mutate(year = ifelse(is.na(year), 2020, year)) %>%
filter(!(name %>% str_detect("Frameset"))) %>%
separate(col = category, into = c("category_1",
"category_2",
"category_3"),
sep = "(?<!\\s)/(?!\\s)") %>%
select(-price_dollar, -description) %>%
mutate(price_euro = as.numeric(price_euro)) %>%
mutate(frame_material = case_when(
name %>% str_detect(" CF ") ~ "carbon",
name %>% str_detect(" CFR ") ~ "carbon",
TRUE ~ "aluminium"
)
) %>%
rename("model" = "name") %>%
mutate(category_1 = replace(category_1,
model == "Speedmax CF SLX 8.0 SL", "Road")) %>%
mutate(category_2 = replace(category_2,
model == "Speedmax CF SLX 8.0 SL", "Triathlon Bike")) %>%
mutate(category_3 = replace(category_3,
model == "Speedmax CF SLX 8.0 SL", "Speedmax")) %>%
left_join(weight_tbl)
# scrape weights ----------------------------------------------------------
for (i in seq_along(bike_data$url_base)) {
html <- read_html(bike_data$url_base[i])
weight <- html %>%
html_nodes(xpath = "//*[@class='eyebrow productDescription__characteristicsLabel'and contains(text(),'Weight')]//following-sibling::div") %>%
html_text() %>% gsub("\\n| |kg", "", .) %>%
as.numeric()
weight_tbl$weight[i] <- weight
Sys.sleep(1)
print(i)
}For each of the 218 models, we know their categories, price and weight. Let’s see how they are related. As a first step in modeling, it’s always a good idea to plot the data:
ggplot(bike_data_tbl,
aes(x = price_euro,
y = weight,
group = category_1,
col = category_1)) +
geom_point() +
geom_smooth(method = lm, se = FALSE) +
scale_color_manual(values=c("#2dc6d6", "#d65a2d", "#d6af2d", "#8a2dd6"))
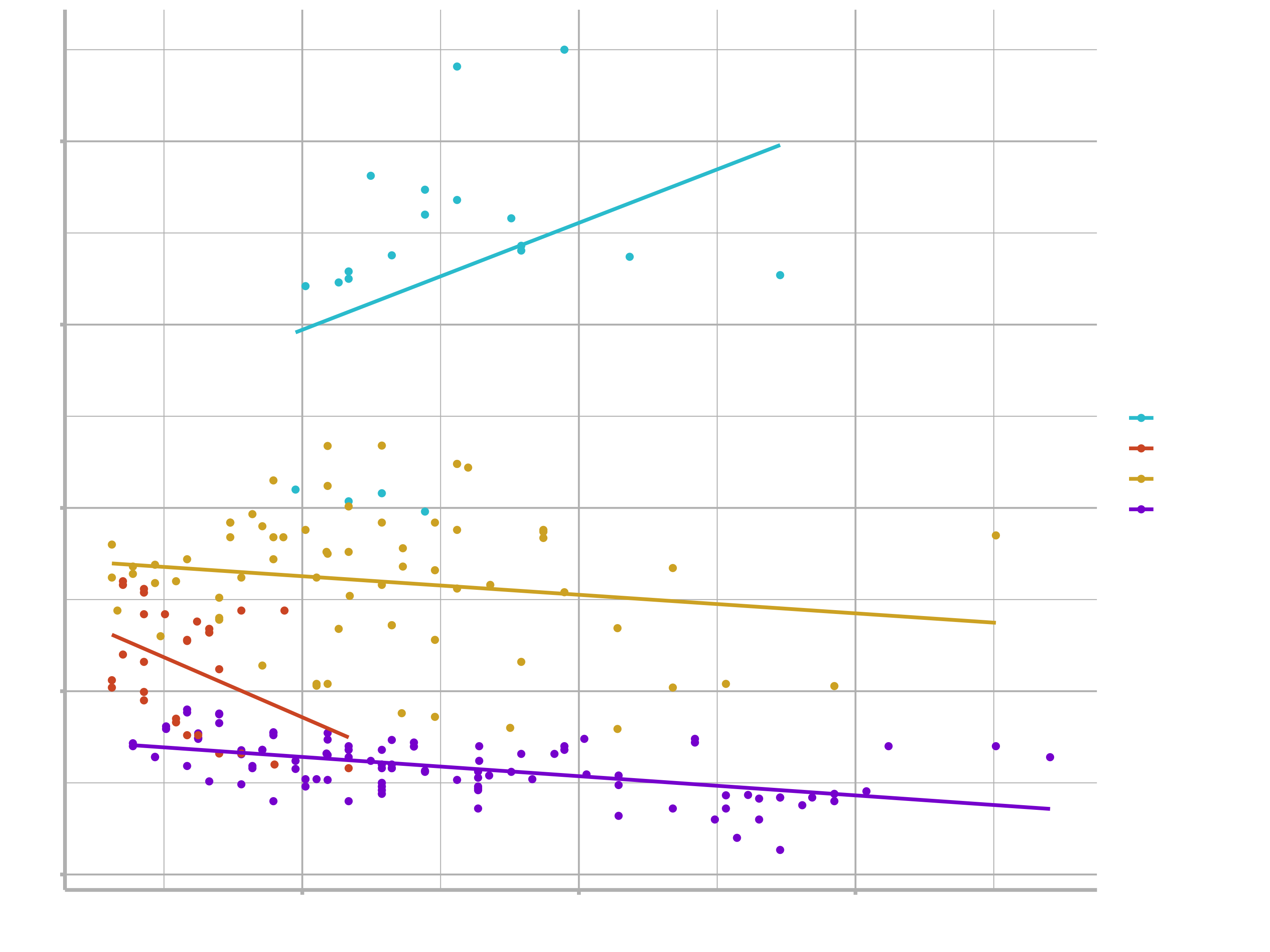
We can see that bikes that are more expensive tend to have lower weights, but the slopes of the lines look different so this effect may depend on the category. Moreover, the relationship behaves exactly the other way for e-bikes.
II. Build and fit a model
A standard two-way analysis of variance (ANOVA) model makes sense for this dataset because we have both a continuous predictor and a categorical predictor. Since the slopes appear to be different for at least some of the bike models, let’s build a model that allows for two-way interactions. Specifying an R formula with our variables in this way:
weight ~ price_euro * category_1
allows our regression model depending on price_euro to have separate slopes and intercepts for each category_1.
For this kind of model, ordinary least squares is a good initial approach. With tidymodels, we start by specifying the functional form of the model that we want using the parsnip package. Since there is a numeric outcome and the model should be linear with slopes and intercepts, the model type is linear regression. We can declare this with:
linear_reg()
## Linear Regression Model Specification (regression)
That is pretty underwhelming since, on its own, it doesn’t really do much. However, now that the type of model has been specified, a method for fitting or training the model can be stated using the engine. The engine value is often a mash-up of the software that can be used to fit or train the model as well as the estimation method. For example, to use ordinary least squares, we can set the engine to be lm:
lm_mod <- linear_reg() %>%
set_engine("lm")
lm_mod
## Linear Regression Model Specification (regression)
##
## Computational engine: lm
From here, the model can be estimated or trained using the fit() function:
lm_fit <- lm_mod %>%
fit(weight ~ price_euro * category_1,
data = bike_data_tbl)
Let’s take a look at the model parameter estimates and their statistical properties with the tidy() function:
tidy(lm_fit)
## # A tibble: 8 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 16.9 1.35 12.6 2.31e-27
## 2 price_euro 0.00117 0.000345 3.38 8.55e- 4
## 3 category_1Hybrid / City -4.38 1.64 -2.68 7.98e- 3
## 4 category_1Mountain -3.30 1.42 -2.33 2.07e- 2
## 5 category_1Road -8.21 1.39 -5.89 1.51e- 8
## 6 price_euro:category_1Hybrid / City -0.00248 0.000698 -3.55 4.76e- 4
## 7 price_euro:category_1Mountain -0.00137 0.000367 -3.74 2.41e- 4
## 8 price_euro:category_1Road -0.00138 0.000355 -3.88 1.39e- 4
III. Use a model to predict
Suppose that it would be particularly interesting to make a plot of the bike weights that cost 2000 €. To create such a graph, we start with some new example data that we will make predictions for, to show in our graph:
new_points <- expand.grid(price_euro = 20000,
category_1 = c("E-Bikes", "Hybrid / City", "Mountain", "Road"))
new_points
## price_euro category_1
## 1 2000 E-Bikes
## 2 2000 Hybrid / City
## 3 2000 Mountain
## 4 2000 Road
To get our predicted results, we can use the predict() function to find the mean values at 2000 €.
First, let’s generate the mean bike weight values:
mean_pred <- predict(lm_fit, new_data = new_points)
mean_pred
## # A tibble: 4 x 1
## .pred
## <dbl>
## 1 19.3
## 2 9.94
## 3 13.2
## 4 8.31
When making predictions, the tidymodels convention is to always produce a tibble of results with standardized column names. This makes it easy to combine the original data and the predictions in a usable format:
conf_int_pred <- predict(lm_fit,
new_data = new_points,
type = "conf_int")
conf_int_pred
## # A tibble: 4 x 2
## .pred_lower .pred_upper
## <dbl> <dbl>
## 1 17.9 20.7
## 2 9.04 10.8
## 3 12.8 13.7
## 4 7.88 8.74
# Now combine:
plot_data <- new_points %>%
bind_cols(mean_pred) %>%
bind_cols(conf_int_pred)
# and plot:
ggplot(plot_data, aes(x = category_1)) +
geom_point(aes(y = .pred)) +
geom_errorbar(aes(ymin = .pred_lower,
ymax = .pred_upper),
width = .2) +
labs(y = "Bike weight", x = "Category")
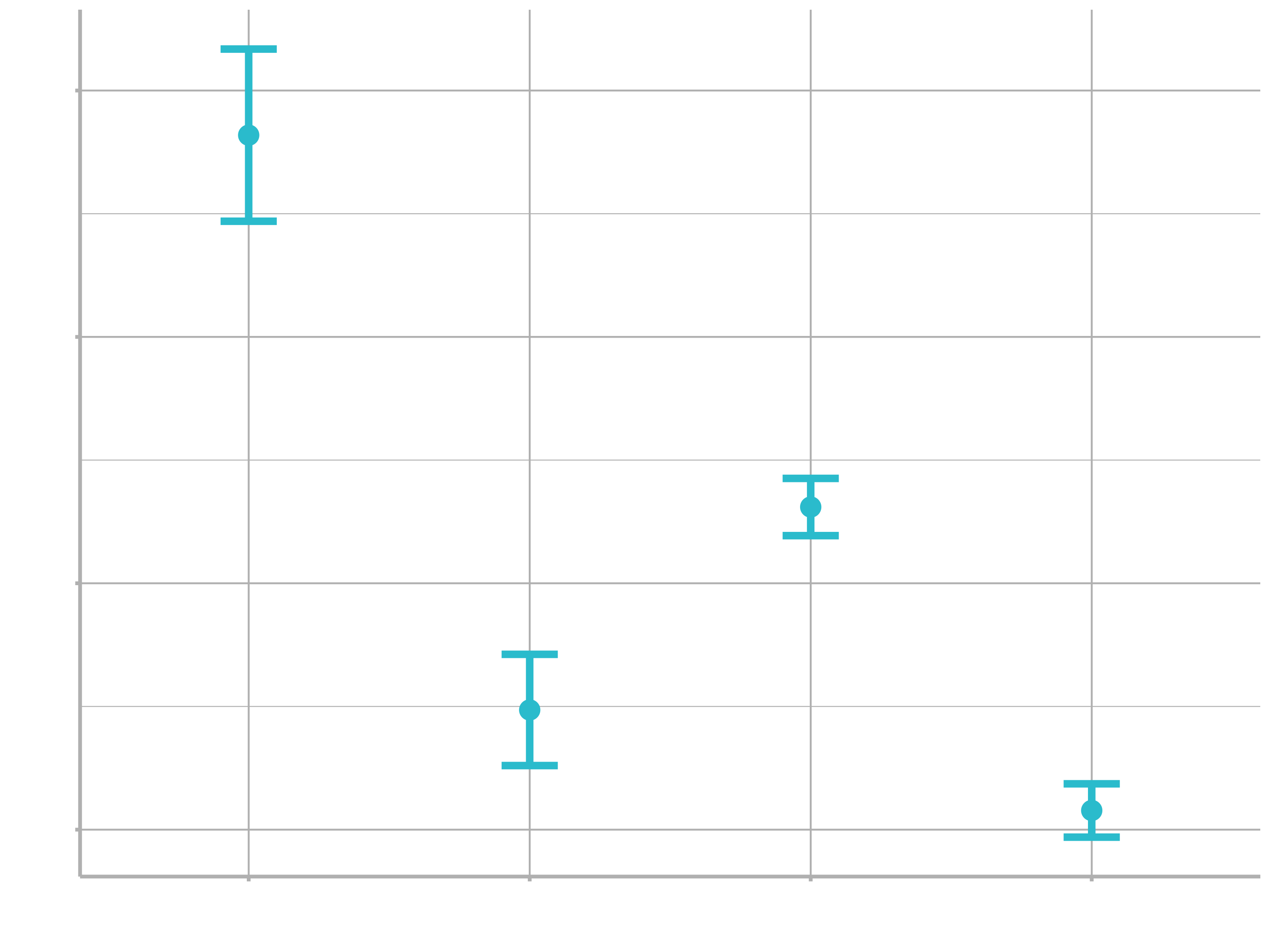
IV. Model with a different engine
Every one on your team is happy with that plot except that one person who just read their first book on Bayesian analysis. They are interested in knowing if the results would be different if the model were estimated using a Bayesian approach. In such an analysis, a prior distribution needs to be declared for each model parameter that represents the possible values of the parameters (before being exposed to the observed data). After some discussion, the group agrees that the priors should be bell-shaped but, since no one has any idea what the range of values should be, to take a conservative approach and make the priors wide using a Cauchy distribution (which is the same as a t-distribution with a single degree of freedom).
The documentation on the rstanarm package shows us that the stan_glm() function can be used to estimate this model, and that the function arguments that need to be specified are called prior and prior_intercept. It turns out that linear_reg() has a has a stan engine. Since these prior distribution arguments are specific to the Stan software, they are passed as arguments to parsnip::set_engine(). After that, the same exact fit() call is used:
# set the prior distribution
prior_dist <- rstanarm::student_t(df = 1)
set.seed(123)
# make the parsnip model
bayes_mod <- linear_reg() %>%
set_engine("stan",
prior_intercept = prior_dist,
prior = prior_dist)
# train the model
bayes_fit <- bayes_mod %>%
fit(weight ~ price_euro * category_1,
data = bike_data_tbl)
print(bayes_fit, digits = 5)
## parsnip model object
##
## Fit time: 1m 39.2s
## stan_glm
## family: gaussian [identity]
## formula: weight ~ price_euro * category_1
## observations: 218
## predictors: 8
## ------
## Median MAD_SD
## (Intercept) 15.24134 1.25450
## price_euro 0.00158 0.00032
## category_1Hybrid / City -2.34716 1.53781
## category_1Mountain -1.52799 1.31226
## category_1Road -6.47532 1.31294
## price_euro:category_1Hybrid / City -0.00310 0.00066
## price_euro:category_1Mountain -0.00180 0.00035
## price_euro:category_1Road -0.00180 0.00033
##
## Auxiliary parameter(s):
## Median MAD_SD
## sigma 1.63413 0.07824
##
## ------
## * For help interpreting the printed output see ?print.stanreg
## * For info on the priors used see ?prior_summary.stanreg
This kind of Bayesian analysis (like many models) involves randomly generated numbers in its fitting procedure. We can use set.seed() to ensure that the same (pseudo-)random numbers are generated each time we run this code. The number 123 isn’t special or related to our data; it is just a “seed” used to choose random numbers.
To update the parameter table, the tidy() method is once again used:
tidy(bayes_fit, conf.int = TRUE)
## # A tibble: 8 x 5
## term estimate std.error conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 15.2 1.25 13.3 17.4
## 2 price_euro 0.00158 0.000324 0.00102 0.00209
## 3 category_1Hybrid / City -2.35 1.54 -5.08 -0.0920
## 4 category_1Mountain -1.53 1.31 -3.89 0.418
## 5 category_1Road -6.48 1.31 -8.78 -4.47
## 6 price_euro:category_1Hybrid / City -0.00310 0.000663 -0.00415 -0.00192
## 7 price_euro:category_1Mountain -0.00180 0.000348 -0.00232 -0.00119
## 8 price_euro:category_1Road -0.00180 0.000333 -0.00233 -0.00121
A goal of the tidymodels packages is that the interfaces to common tasks are standardized (as seen in the tidy() results above). The same is true for getting predictions; we can use the same code even though the underlying packages use very different syntax:
bayes_plot_data <-
new_points %>% 0
bind_cols(predict(bayes_fit, new_data = new_points)) %>%
bind_cols(predict(bayes_fit, new_data = new_points, type = "conf_int"))
ggplot(bayes_plot_data, aes(x = food_regime)) +
geom_point(aes(y = .pred)) +
geom_errorbar(aes(ymin = .pred_lower, ymax = .pred_upper), width = .2) +
labs(y = "Bike weight") +
ggtitle("Bayesian model with t(1) prior distribution")
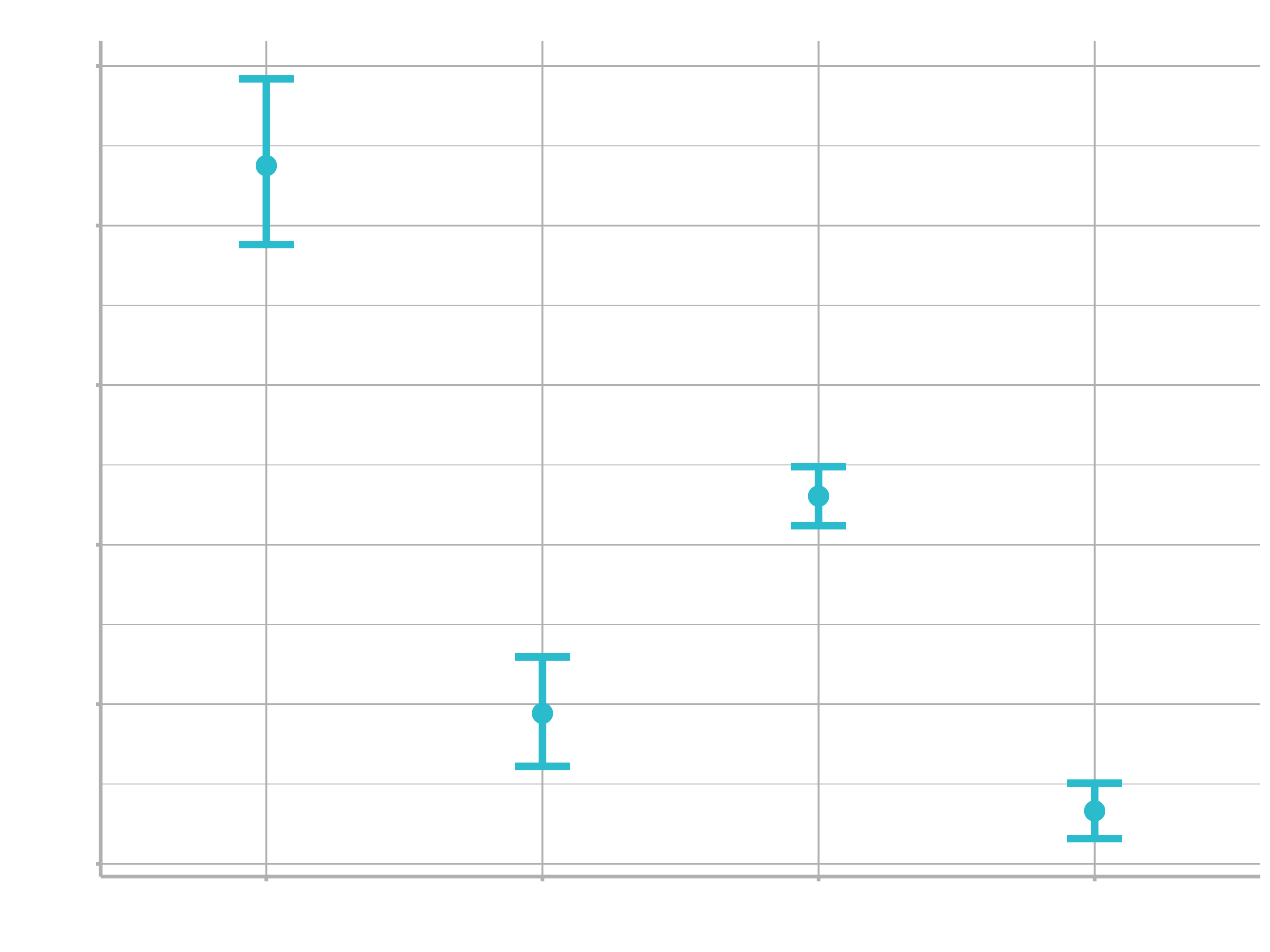
This isn’t very different from the non-Bayesian results (except in interpretation).
2. Preprocessing
Now we have learned how to specify and train models with different engines using the parsnip package. In this stepp, we’ll explore another tidymodels package, recipes, which is designed to help you preprocess your data before training your model. Recipes are built as a series of preprocessing steps, such as:
- converting qualitative predictors to indicator variables (also known as dummy variables),
- transforming data to be on a different scale (e.g., taking the logarithm of a variable),
- transforming whole groups of predictors together,
- extracting key features from raw variables (e.g., getting the day of the week out of a date variable),
and so on.
To use code in this article, you will need the following packages: nycflights13 and tidymodels (and optionally skimr).
I. The data
Let’s use the nycflights13 data to predict whether a plane arrives more than 30 minutes late. This data set contains information on 325,819 flights departing near New York City in 2013. Let’s start by loading the data and making a few changes to the variables:
set.seed(123)
flight_data <-
flights %>%
mutate(
# Convert the arrival delay to a factor
arr_delay = ifelse(arr_delay >= 30, "late", "on_time"),
arr_delay = factor(arr_delay),
# We will use the date (not date-time) in the recipe below
date = as.Date(time_hour)
) %>%
# Include the weather data
inner_join(weather, by = c("origin", "time_hour")) %>%
# Only retain the specific columns we will use
select(dep_time, flight, origin, dest, air_time, distance,
carrier, date, arr_delay, time_hour) %>%
# Exclude missing data
na.omit() %>%
# For creating models, it is better to have qualitative columns
# encoded as factors (instead of character strings)
mutate_if(is.character, as.factor)
We can see that about 16% of the flights in this data set arrived more than 30 minutes late.
flight_data %>%
count(arr_delay) %>%
mutate(prop = n/sum(n))
## # A tibble: 2 x 3
## arr_delay n prop
## <fct> <int> <dbl>
## 1 late 52540 0.161
## 2 on_time 273279 0.839
Before we start building up our recipe, let’s take a quick look at a few specific variables that will be important for both preprocessing and modeling.
First, notice that the variable we created called arr_delay is a factor variable; it is important that our outcome variable for training a logistic regression model is a factor.
glimpse(flight_data)
## Rows: 325,819
## Columns: 10
## $ dep_time <int> 517, 533, 542, 544, 554, 554, 555, 557, 557, 558, 558, 558,…
## $ flight <int> 1545, 1714, 1141, 725, 461, 1696, 507, 5708, 79, 301, 49, 7…
## $ origin <fct> EWR, LGA, JFK, JFK, LGA, EWR, EWR, LGA, JFK, LGA, JFK, JFK,…
## $ dest <fct> IAH, IAH, MIA, BQN, ATL, ORD, FLL, IAD, MCO, ORD, PBI, TPA,…
## $ air_time <dbl> 227, 227, 160, 183, 116, 150, 158, 53, 140, 138, 149, 158, …
## $ distance <dbl> 1400, 1416, 1089, 1576, 762, 719, 1065, 229, 944, 733, 1028…
## $ carrier <fct> UA, UA, AA, B6, DL, UA, B6, EV, B6, AA, B6, B6, UA, UA, AA,…
## $ date <date> 2013-01-01, 2013-01-01, 2013-01-01, 2013-01-01, 2013-01-01…
## $ arr_delay <fct> on_time, on_time, late, on_time, on_time, on_time, on_time,…
## $ time_hour <dttm> 2013-01-01 05:00:00, 2013-01-01 05:00:00, 2013-01-01 05:00…
Second, there are two variables that we don’t want to use as predictors in our model, but that we would like to retain as identification variables that can be used to troubleshoot poorly predicted data points. These are flight, a numeric value, and time_hour, a date-time value.
Third, there are 104 flight destinations contained in dest and 16 distinct carriers.
flight_data %>%
skimr::skim(dest, carrier)
── Data Summary ────────────────────────
Values
Name Piped data
Number of rows 325819
Number of columns 10
_______________________
Column type frequency:
factor 2
________________________
Group variables None
── Variable type: factor ──────────────────────────────────────────────────────
skim_variable n_missing complete_rate ordered n_unique top_counts
1 dest 0 1 FALSE 104 ATL: 16771, ORD: 16507
2 carrier 0 1 FALSE 16 UA: 57489, B6: 53715
Because we’ll be using a simple logistic regression model, the variables dest and carrier will be converted to dummy variables. However, some of these values do not occur very frequently and this could complicate our analysis. We’ll discuss specific steps later in this session that we can add to our recipe to address this issue before modeling.
II. Data Splitting
To get started, let’s split this single dataset into two: a training set and a testing set. We’ll keep most of the rows in the original dataset (subset chosen randomly) in the training set. The training data will be used to fit the model, and the testing set will be used to measure model performance.
To do this, we can use the rsample package to create an object that contains the information on how to split the data, and then two more rsample functions to create data frames for the training and testing sets:
# Fix the random numbers by setting the seed
# This enables the analysis to be reproducible when random numbers are used
set.seed(555)
# Put 3/4 of the data into the training set
data_split <- initial_split(flight_data, prop = 3/4)
# Create data frames for the two sets:
train_data <- training(data_split)
test_data <- testing(data_split)
III. Create recipe and roles
To get started, let’s create a recipe for a simple logistic regression model. Before training the model, we can use a recipe to create a few new predictors and conduct some preprocessing required by the model.
Let’s initiate a new recipe:
flights_rec <-
recipe(arr_delay ~ ., data = train_data)
The recipe() function as we used it here has two arguments:
- A formula. Any variable on the left-hand side of the tilde (
~) is considered the model outcome (here,arr_delay). On the right-hand side of the tilde are the predictors. Variables may be listed by name, or you can use the dot (.) to indicate all other variables as predictors. - The data. A recipe is associated with the data set used to create the model. This will typically be the training set, so
data = train_datahere. Naming a data set doesn’t actually change the data itself; it is only used to catalog the names of the variables and their types, like factors, integers, dates, etc.
Now we can add roles to this recipe. We can use the update_role() function to let recipes know that flight and time_hour are variables with a custom role that we called "ID" (a role can have any character value). Whereas our formula included all variables in the training set other than arr_delay as predictors, this tells the recipe to keep these two variables but not use them as either outcomes or predictors.
flights_rec <-
recipe(arr_delay ~ ., data = train_data) %>%
update_role(flight, time_hour, new_role = "ID")
This step of adding roles to a recipe is optional; the purpose of using it here is that those two variables can be retained in the data but not included in the model. This can be convenient when, after the model is fit, we want to investigate some poorly predicted value. These ID columns will be available and can be used to try to understand what went wrong.
To get the current set of variables and roles, use the summary() function:
summary(flights_rec)
## # A tibble: 10 x 4
## variable type role source
## <chr> <chr> <chr> <chr>
## 1 dep_time numeric predictor original
## 2 flight numeric ID original
## 3 origin nominal predictor original
## 4 dest nominal predictor original
## 5 air_time numeric predictor original
## 6 distance numeric predictor original
## 7 carrier nominal predictor original
## 8 date date predictor original
## 9 time_hour date ID original
## 10 arr_delay nominal outcome original
IV. Create features
Now we can start adding steps onto our recipe using the pipe operator. Perhaps it is reasonable for the date of the flight to have an effect on the likelihood of a late arrival. A little bit of feature engineering might go a long way to improving our model. How should the date be encoded into the model? The date column has an R date object so including that column “as is” will mean that the model will convert it to a numeric format equal to the number of days after a reference date:
flight_data %>%
distinct(date) %>%
mutate(numeric_date = as.numeric(date))
## # A tibble: 364 x 2
## date numeric_date
## <date> <dbl>
## 1 2013-01-01 15706
## 2 2013-01-02 15707
## 3 2013-01-03 15708
## 4 2013-01-04 15709
## 5 2013-01-05 15710
## # … with 359 more rows
It’s possible that the numeric date variable is a good option for modeling; perhaps the model would benefit from a linear trend between the log-odds of a late arrival and the numeric date variable. However, it might be better to add model terms derived from the date that have a better potential to be important to the model. For example, we could derive the following meaningful features from the single date variable:
- the day of the week,
- the month, and
- whether or not the date corresponds to a holiday.
Let’s do all three of these by adding steps to our recipe:
flights_rec <-
recipe(arr_delay ~ ., data = train_data) %>%
update_role(flight, time_hour, new_role = "ID") %>%
step_date(date, features = c("dow", "month")) %>%
step_holiday(date, holidays = timeDate::listHolidays("US")) %>%
step_rm(date)
What do each of these steps do?
- With
step_date(), we created two new factor columns with the appropriate day of the week and the month. - With
step_holiday(), we created a binary variable indicating whether the current date is a holiday or not. The argument value of timeDate::listHolidays(“US”) uses thetimeDatepackage to list the 17 standard US holidays. - With
step_rm(), we remove the original date variable since we no longer want it in the model.
Next, we’ll turn our attention to the variable types of our predictors. Because we plan to train a logistic regression model, we know that predictors will ultimately need to be numeric, as opposed to factor variables. In other words, there may be a difference in how we store our data (in factors inside a data frame), and how the underlying equations require them (a purely numeric matrix).
For factors like dest and origin, standard practice is to convert them into dummy or indicator variables to make them numeric. These are binary values for each level of the factor. For example, our origin variable has values of "EWR", "JFK", and "LGA". The standard dummy variable encoding, shown below, will create two numeric columns of the data that are 1 when the originating airport is "JFK" or "LGA" and zero otherwise, respectively.
| ORIGIN | ORIGIN_JFK | ORIGIN_LGA |
|---|---|---|
| EWR | 0 | 0 |
| JFK | 1 | 0 |
| LGA | 0 | 1 |
But, unlike the standard model formula methods in R, a recipe does not automatically create these dummy variables for you; you’ll need to tell your recipe to add this step. This is for two reasons. First, many models do not require numeric predictors, so dummy variables may not always be preferred. Second, recipes can also be used for purposes outside of modeling, where non-dummy versions of the variables may work better. For example, you may want to make a table or a plot with a variable as a single factor. For those reasons, you need to explicitly tell recipes to create dummy variables using step_dummy():
flights_rec <-
recipe(arr_delay ~ ., data = train_data) %>%
update_role(flight, time_hour, new_role = "ID") %>%
step_date(date, features = c("dow", "month")) %>%
step_holiday(date, holidays = timeDate::listHolidays("US")) %>%
step_rm(date) %>%
step_dummy(all_nominal(), -all_outcomes())
Here, we did something different than before: instead of applying a step to an individual variable, we used selectors to apply this recipe step to several variables at once.
- The first selector,
all_nominal(), selects all variables that are either factors or characters. - The second selector,
-all_outcomes()removes any outcome variables from this recipe step. With these two selectors together, our recipe step above translates to:
Create dummy variables for all of the factor or character columns unless they are outcomes.
At this stage in the recipe, this step selects the origin, dest, and carrier variables. It also includes two new variables, date_dow (day of week) and date_month, that were created by the earlier step_date().
More generally, the recipe selectors mean that you don’t always have to apply steps to individual variables one at a time. Since a recipe knows the variable type and role of each column, they can also be selected (or dropped) using this information.
We need one final step to add to our recipe. Since carrier and dest have some infrequently occurring factor values, it is possible that dummy variables might be created for values that don’t exist in the training set. For example, there is one destination that is only in the test set:
test_data %>%
distinct(dest) %>%
anti_join(train_data)
## Joining, by = "dest"
## # A tibble: 1 x 1
## dest
## <fct>
## 1 LEX
When the recipe is applied to the training set, a column is made for LEX because the factor levels come from flight_data (not the training set), but this column will contain all zeros. This is a “zero-variance predictor” that has no information within the column. While some R functions will not produce an error for such predictors, it usually causes warnings and other issues. step_zv() will remove columns from the data when the training set data have a single value, so it is added to the recipe after step_dummy():
flights_rec <-
recipe(arr_delay ~ ., data = train_data) %>%
update_role(flight, time_hour, new_role = "ID") %>%
step_date(date, features = c("dow", "month")) %>%
step_holiday(date, holidays = timeDate::listHolidays("US")) %>%
step_rm(date) %>%
step_dummy(all_nominal(), -all_outcomes()) %>%
step_zv(all_predictors())
Now we’ve created a specification of what should be done with the data. How do we use the recipe we made?
V. Fit a model with a recipe
Let’s use logistic regression to model the flight data. As we saw in 1. Modeling, we start by building a model specification using the parsnip package:
lr_mod <-
logistic_reg() %>%
set_engine("glm")
We will want to use our recipe across several steps as we train and test our model. We will:
- Process the recipe using the training set: This involves any estimation or calculations based on the training set. For our recipe, the training set will be used to determine which predictors should be converted to dummy variables and which predictors will have zero-variance in the training set, and should be slated for removal.
- Apply the recipe to the training set: We create the final predictor set on the training set.
- Apply the recipe to the test set: We create the final predictor set on the test set. Nothing is recomputed and no information from the test set is used here; the dummy variable and zero-variance results from the training set are applied to the test set.
To simplify this process, we can use a model workflow, which pairs a model and recipe together. This is a straightforward approach because different recipes are often needed for different models, so when a model and recipe are bundled, it becomes easier to train and test workflows. We’ll use the workflows package from tidymodels to bundle our parsnip model (lr_mod) with our recipe (flights_rec).
flights_wflow <-
workflow() %>%
add_model(lr_mod) %>%
add_recipe(flights_rec)
flights_wflow
## ══ Workflow ══════════════════════════════════════════════════════════
## Preprocessor: Recipe
## Model: logistic_reg()
##
## ── Preprocessor ──────────────────────────────────────────────────────
## 5 Recipe Steps
##
## ● step_date()
## ● step_holiday()
## ● step_rm()
## ● step_dummy()
## ● step_zv()
##
## ── Model ─────────────────────────────────────────────────────────────
## Logistic Regression Model Specification (classification)
##
## Computational engine: glm
Now, there is a single function that can be used to prepare the recipe and train the model from the resulting predictors:
flights_fit <-
flights_wflow %>%
fit(data = train_data)
This object has the finalized recipe and fitted model objects inside. You may want to extract the model or recipe objects from the workflow. To do this, you can use the helper functions pull_workflow_fit() and pull_workflow_prepped_recipe(). For example, here we pull the fitted model object then use the broom::tidy() function to get a tidy tibble of model coefficients:
flights_fit %>%
pull_workflow_fit() %>%
tidy()
## # A tibble: 157 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 3.91 2.73 1.43 1.51e- 1
## 2 dep_time -0.00167 0.0000141 -118. 0.
## 3 air_time -0.0439 0.000561 -78.4 0.
## 4 distance 0.00686 0.00150 4.57 4.84e- 6
## 5 date_USChristmasDay 1.12 0.173 6.49 8.45e-11
## # … with 152 more rows
VI. Use a trained workflow to predict
Our goal was to predict whether a plane arrives more than 30 minutes late. We have just:
- Built the model (
lr_mod), - Created a preprocessing recipe (
flights_rec), - Bundled the model and recipe (
flights_wflow), and - Trained our workflow using a single call to
fit().
The next step is to use the trained workflow (flights_fit) to predict with the unseen test data, which we will do with a single call to predict(). The predict() method applies the recipe to the new data, then passes them to the fitted model.
predict(flights_fit, test_data)
## # A tibble: 81,454 x 1
## .pred_class
## <fct>
## 1 on_time
## 2 on_time
## 3 on_time
## 4 on_time
## 5 on_time
## # … with 81,449 more rows
Because our outcome variable here is a factor, the output from predict() returns the predicted class: late versus on_time. But, let’s say we want the predicted class probabilities for each flight instead. To return those, we can specify type = "prob" when we use predict(). We’ll also bind the output with some variables from the test data and save them together:
flights_pred <-
predict(flights_fit, test_data, type = "prob") %>%
bind_cols(test_data %>% select(arr_delay, time_hour, flight))
# The data look like:
flights_pred
## # A tibble: 81,454 x 5
## .pred_late .pred_on_time arr_delay time_hour flight
## <dbl> <dbl> <fct> <dttm> <int>
## 1 0.0565 0.944 on_time 2013-01-01 05:00:00 1714
## 2 0.0264 0.974 on_time 2013-01-01 06:00:00 79
## 3 0.0481 0.952 on_time 2013-01-01 06:00:00 301
## 4 0.0325 0.967 on_time 2013-01-01 06:00:00 49
## 5 0.0711 0.929 on_time 2013-01-01 06:00:00 1187
## # … with 81,449 more rows
Now that we have a tibble with our predicted class probabilities, how will we evaluate the performance of our workflow? We can see from these first few rows that our model predicted these 5 on time flights correctly because the values of .pred_on_time are p > .50. But we also know that we have 81,454 rows total to predict. We would like to calculate a metric that tells how well our model predicted late arrivals, compared to the true status of our outcome variable, arr_delay.
Let’s use the area under the ROC curve as our metric, computed using roc_curve() and roc_auc() from the yardstick package.
To generate a ROC curve, we need the predicted class probabilities for late and on_time, which we just calculated in the code chunk above. We can create the ROC curve with these values, using roc_curve() and then piping to the autoplot() method:
flights_pred %>%
roc_curve(truth = arr_delay, .pred_late) %>%
autoplot()
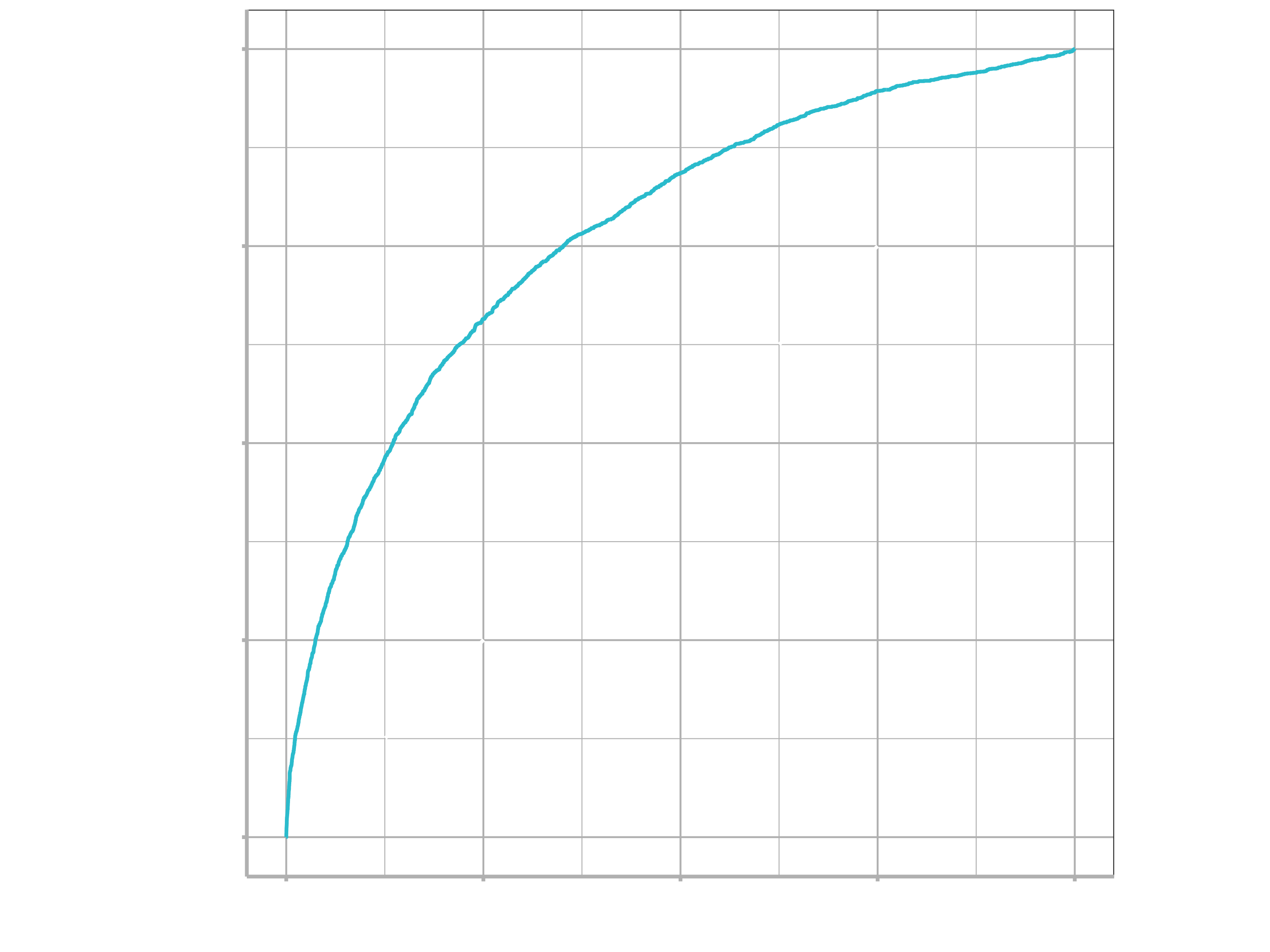
Similarly, roc_auc() estimates the area under the curve:
flights_pred %>%
roc_auc(truth = arr_delay, .pred_late)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 roc_auc binary 0.765
Not too bad! I leave it to you to test out this workflow without this recipe. You can use workflows::add_formula(arr_delay ~ .) instead of add_recipe() (remember to remove the identification variables first!), and see whether our recipe improved our model’s ability to predict late arrivals.
3. Evaluating
So far, we have built a model and preprocessed data with a recipe. We also introduced workflows as a way to bundle a parsnip model and recipe together. Once we have a model trained, we need a way to measure how well that model predicts new data. This section explains how to characterize model performance based on resampling statistics.
Load necessary packages:
library(tidymodels) # for the rsample package, along with the rest of tidymodels
# Helper packages
library(modeldata) # for the cells data
I. The data
Let’s use data from Hill, LaPan, Li, and Haney (2007), available in the modeldata package, to predict cell image segmentation quality with resampling. To start, we load this data into R:
data(cells, package = "modeldata")
cells
We have data for 2019 cells, with 58 variables. The main outcome variable of interest for us here is called class, which you can see is a factor. But before we jump into predicting the class variable, we need to understand it better. Below is a brief primer on cell image segmentation.
Some biologists conduct experiments on cells. In drug discovery, a particular type of cell can be treated with either a drug or control and then observed to see what the effect is (if any). A common approach for this kind of measurement is cell imaging. Different parts of the cells can be colored so that the locations of a cell can be determined.
For example, in top panel of this image of five cells, the green color is meant to define the boundary of the cell (coloring something called the cytoskeleton) while the blue color defines the nucleus of the cell.
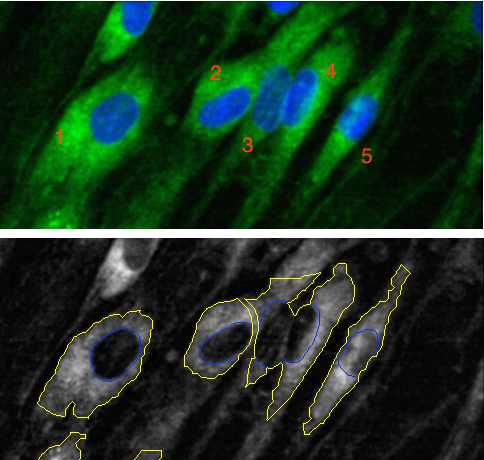
Using these colors, the cells in an image can be segmented so that we know which pixels belong to which cell. If this is done well, the cell can be measured in different ways that are important to the biology. Sometimes the shape of the cell matters and different mathematical tools are used to summarize characteristics like the size or “oblongness” of the cell.
The bottom panel shows some segmentation results. Cells 1 and 5 are fairly well segmented. However, cells 2 to 4 are bunched up together because the segmentation was not very good. The consequence of bad segmentation is data contamination; when the biologist analyzes the shape or size of these cells, the data are inaccurate and could lead to the wrong conclusion.
A cell-based experiment might involve millions of cells so it is unfeasible to visually assess them all. Instead, a subsample can be created and these cells can be manually labeled by experts as either poorly segmented (PS) or well-segmented (WS). If we can predict these labels accurately, the larger data set can be improved by filtering out the cells most likely to be poorly segmented.
The cells data has class labels for 2019 cells — each cell is labeled as either poorly segmented (PS) or well-segmented (WS). Each also has a total of 56 predictors based on automated image analysis measurements. For example, avg_inten_ch_1 is the mean intensity of the data contained in the nucleus, area_ch_1 is the total size of the cell, and so on (some predictors are fairly arcane in nature).
The rates of the classes are somewhat imbalanced; there are more poorly segmented cells than well-segmented cells. Look at proportion of classes:
cells %>%
count(class) %>%
mutate(prop = n/sum(n))
#> # A tibble: 2 x 3
## class n prop
## <fct> <int> <dbl>
## 1 PS 1300 0.644
## 2 WS 719 0.356
II. Data splitting
In the previous section (Preprocessing data with recipes), we started by splitting our data. It is common when beginning a modeling project to separate the data set into two partitions:
- The training set is used to estimate parameters, compare models and feature engineering techniques, tune models, etc.
- The test set is held in reserve until the end of the project, at which point there should only be one or two models under serious consideration. It is used as an unbiased source for measuring final model performance.
There are different ways to create these partitions of the data. The most common approach is to use a random sample. Suppose that one quarter of the data were reserved for the test set. Random sampling would randomly select 25% for the test set and use the remainder for the training set. We can use the rsample package for this purpose.
Since random sampling uses random numbers, it is important to set the random number seed. This ensures that the random numbers can be reproduced at a later time (if needed).
The function rsample::initial_split() takes the original data and saves the information on how to make the partitions. In the original analysis, the authors made their own training/test set and that information is contained in the column case. To demonstrate how to make a split, we’ll remove this column before we make our own split:
set.seed(123)
cell_split <- initial_split(cells %>% select(-case),
strata = class)
Here we used the strata argument, which conducts a stratified split. This ensures that, despite the imbalance we noticed in our class variable, our training and test data sets will keep roughly the same proportions of poorly and well-segmented cells as in the original data. After the initial_split, the training() and testing() functions return the actual data sets.
cell_train <- training(cell_split)
cell_test <- testing(cell_split)
nrow(cell_train)
## 1515
nrow(cell_train)/nrow(cells)
## 0.7503715
# training set proportions by class
cell_train %>%
count(class) %>%
mutate(prop = n/sum(n))
## # A tibble: 2 x 3
## class n prop
## <fct> <int> <dbl>
## 1 PS 975 0.644
## 2 WS 540 0.356
# test set proportions by class
cell_test %>%
count(class) %>%
mutate(prop = n/sum(n))
## # A tibble: 2 x 3
## class n prop
## <fct> <int> <dbl>
## 1 PS 325 0.645
## 2 WS 179 0.355
The majority of the modeling work is then conducted on the training set data.
III. Modeling
Random forest models are ensembles of decision trees. A large number of decision tree models are created for the ensemble based on slightly different versions of the training set. When creating the individual decision trees, the fitting process encourages them to be as diverse as possible. The collection of trees are combined into the random forest model and, when a new sample is predicted, the votes from each tree are used to calculate the final predicted value for the new sample. For categorical outcome variables like class in our cells data example, the majority vote across all the trees in the random forest determines the predicted class for the new sample.
One of the benefits of a random forest model is that it is very low maintenance; it requires very little preprocessing of the data and the default parameters tend to give reasonable results. For that reason, we won’t create a recipe for the cells data.
At the same time, the number of trees in the ensemble should be large (in the thousands) and this makes the model moderately expensive to compute.
To fit a random forest model on the training set, let’s use the parsnip package with the ranger engine. We first define the model that we want to create:
rf_mod <-
rand_forest(trees = 1000) %>%
set_engine("ranger") %>%
set_mode("classification")
Starting with this parsnip model object, the fit() function can be used with a model formula. Since random forest models use random numbers, we again set the seed prior to computing:
set.seed(234)
rf_fit <-
rf_mod %>%
fit(class ~ ., data = cell_train)
rf_fit
## parsnip model object
##
## Fit time: 2.4s
## Ranger result
##
## Call:
## ranger::ranger(formula = class ~ ., data = data, num.trees = ~1000, num.threads = 1, verbose = FALSE, seed = sample.int(10^5, 1), probability = TRUE)
##
## Type: Probability estimation
## Number of trees: 1000
## Sample size: 1515
## Number of independent variables: 56
## Mtry: 7
## Target node size: 10
## Variable importance mode: none
## Splitrule: gini
## OOB prediction error (Brier s.): 0.1218873
This new rf_fit object is our fitted model, trained on our training data set.
IV. Estimating performance
During a modeling project, we might create a variety of different models. To choose between them, we need to consider how well these models do, as measured by some performance statistics. In our example in this article, some options we could use are:
- the area under the Receiver Operating Characteristic (ROC) curve, and
- overall classification accuracy.
The ROC curve uses the class probability estimates to give us a sense of performance across the entire set of potential probability cutoffs. Overall accuracy uses the hard class predictions to measure performance. The hard class predictions tell us whether our model predicted PS or WS for each cell. But, behind those predictions, the model is actually estimating a probability. A simple 50% probability cutoff is used to categorize a cell as poorly segmented.
The yardstick package has functions for computing both of these measures called roc_auc() and accuracy().
At first glance, it might seem like a good idea to use the training set data to compute these statistics. (This is actually a very bad idea.) Let’s see what happens if we try this. To evaluate performance based on the training set, we call the predict() method to get both types of predictions (i.e. probabilities and hard class predictions).
rf_training_pred <-
predict(rf_fit, cell_train) %>%
bind_cols(predict(rf_fit, cell_train, type = "prob")) %>%
# Add the true outcome data back in
bind_cols(cell_train %>%
select(class))
Using the yardstick functions, this model has spectacular results, so spectacular that you might be starting to get suspicious:
rf_training_pred %>% # training set predictions
roc_auc(truth = class, .pred_PS)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 roc_auc binary 1.00
rf_training_pred %>% # training set predictions
accuracy(truth = class, .pred_class)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.993
Now that we have this model with exceptional performance, we proceed to the test set. Unfortunately, we discover that, although our results aren’t bad, they are certainly worse than what we initially thought based on predicting the training set:
rf_testing_pred <-
predict(rf_fit, cell_test) %>%
bind_cols(predict(rf_fit, cell_test, type = "prob")) %>%
bind_cols(cell_test %>% select(class))
And look at performance results from prediction with the test set:
rf_testing_pred %>% # test set predictions
roc_auc(truth = class, .pred_PS)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 roc_auc binary 0.909
rf_testing_pred %>% # test set predictions
accuracy(truth = class, .pred_class)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.837
Whoops! Our performance results with the training set were a little too good to be true!
There are several reasons why training set statistics like the ones shown in this section can be unrealistically optimistic:
- Models like random forests, neural networks, and other black-box methods can essentially memorize the training set. Re-predicting that same set should always result in nearly perfect results.
- The training set does not have the capacity to be a good arbiter of performance. It is not an independent piece of information; predicting the training set can only reflect what the model already knows.
To understand that second point better, think about an analogy from teaching. Suppose you give a class a test, then give them the answers, then provide the same test. The student scores on the second test do not accurately reflect what they know about the subject; these scores would probably be higher than their results on the first test.
V. Resampling to the rescue
Resampling methods, such as cross-validation and the bootstrap, are empirical simulation systems. They create a series of data sets similar to the training/testing split discussed previously; a subset of the data are used for creating the model and a different subset is used to measure performance. Resampling is always used with the training set. This schematic from Kuhn and Johnson (2019) illustrates data usage for resampling methods:
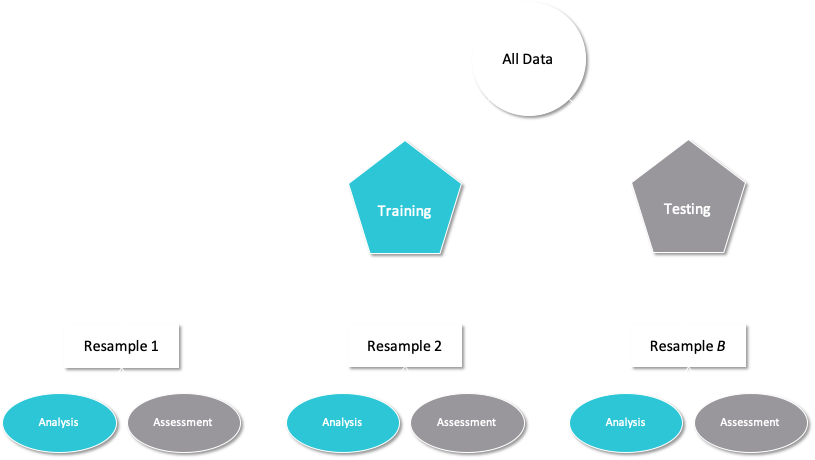
In the first level of this diagram, you see what happens when you use rsample::initial_split(), which splits the original data into training and test sets. Then, the training set is chosen for resampling, and the test set is held out.
Let’s use 10-fold cross-validation (CV) in this example. This method randomly allocates the 1515 cells in the training set to 10 groups of roughly equal size, called “folds”. For the first iteration of resampling, the first fold of about 151 cells are held out for the purpose of measuring performance. This is similar to a test set but, to avoid confusion, we call these data the assessment set in the tidymodels framework.
The other 90% of the data (about 1363 cells) are used to fit the model. Again, this sounds similar to a training set, so in tidymodels we call this data the analysis set. This model, trained on the analysis set, is applied to the assessment set to generate predictions, and performance statistics are computed based on those predictions.
In this example, 10-fold CV moves iteratively through the folds and leaves a different 10% out each time for model assessment. At the end of this process, there are 10 sets of performance statistics that were created on 10 data sets that were not used in the modeling process. For the cell example, this means 10 accuracies and 10 areas under the ROC curve. While 10 models were created, these are not used further; we do not keep the models themselves trained on these folds because their only purpose is calculating performance metrics.
The final resampling estimates for the model are the averages of the performance statistics replicates. For example, suppose for our data the results were:
| RESAMPLE | ACCURACY | ROC_AUC | ASSESSMENT SIZE |
|---|---|---|---|
| Fold01 | 0.7828947 | 0.8419206 | 152 |
| Fold02 | 0.8092105 | 0.8939982 | 152 |
| Fold03 | 0.8486842 | 0.9174923 | 152 |
| Fold04 | 0.8355263 | 0.8941946 | 152 |
| Fold05 | 0.8684211 | 0.9063232 | 152 |
| Fold06 | 0.8410596 | 0.9136661 | 151 |
| Fold07 | 0.8807947 | 0.9368932 | 151 |
| Fold08 | 0.7814570 | 0.8890798 | 151 |
| Fold09 | 0.8145695 | 0.9075369 | 151 |
| Fold10 | 0.8675497 | 0.9310806 | 151 |
From these resampling statistics, the final estimate of performance for this random forest model would be 0.903 for the area under the ROC curve and 0.833 for accuracy.
These resampling statistics are an effective method for measuring model performance without predicting the training set directly as a whole.
VI. Fit a model with resampling
To generate these results, the first step is to create a resampling object using rsample. There are several resampling methods implemented in rsample; cross-validation folds can be created using vfold_cv():
set.seed(345)
folds <- vfold_cv(cell_train, v = 10)
folds
## # 10-fold cross-validation
## # A tibble: 10 x 2
## splits id
## <list> <chr>
## 1 <split [1.4K/152]> Fold01
## 2 <split [1.4K/152]> Fold02
## 3 <split [1.4K/152]> Fold03
## 4 <split [1.4K/152]> Fold04
## 5 <split [1.4K/152]> Fold05
## 6 <split [1.4K/151]> Fold06
## 7 <split [1.4K/151]> Fold07
## 8 <split [1.4K/151]> Fold08
## 9 <split [1.4K/151]> Fold09
## 10 <split [1.4K/151]> Fold10
The list column for splits contains the information on which rows belong in the analysis and assessment sets. There are functions that can be used to extract the individual resampled data called analysis() and assessment().
However, the tune package contains high-level functions that can do the required computations to resample a model for the purpose of measuring performance. You have several options for building an object for resampling:
Resample a model specification preprocessed with a formula or
recipe, orResample a
workflow()that bundles together a model specification and formula/recipe.
For this example, let’s use a workflow() that bundles together the random forest model and a formula, since we are not using a recipe. Whichever of these options you use, the syntax to fit_resamples() is very similar to fit():
rf_wf <-
workflow() %>%
add_model(rf_mod) %>%
add_formula(class ~ .)
Now apply the workflow and fit the model with each fold:
(This computation will take a bit, so be patient.)
set.seed(456)
rf_fit_rs <-
rf_wf %>%
fit_resamples(folds)
rf_fit_rs
## # Resampling results
## # 10-fold cross-validation
## # A tibble: 10 x 4
## splits id .metrics .notes
## <list> <chr> <list> <list>
## 1 <split [1.4K/152]> Fold01 <tibble [2 × 3]> <tibble [0 × 1]>
## 2 <split [1.4K/152]> Fold02 <tibble [2 × 3]> <tibble [0 × 1]>
## 3 <split [1.4K/152]> Fold03 <tibble [2 × 3]> <tibble [0 × 1]>
## 4 <split [1.4K/152]> Fold04 <tibble [2 × 3]> <tibble [0 × 1]>
## 5 <split [1.4K/152]> Fold05 <tibble [2 × 3]> <tibble [0 × 1]>
## 6 <split [1.4K/151]> Fold06 <tibble [2 × 3]> <tibble [0 × 1]>
## 7 <split [1.4K/151]> Fold07 <tibble [2 × 3]> <tibble [0 × 1]>
## 8 <split [1.4K/151]> Fold08 <tibble [2 × 3]> <tibble [0 × 1]>
## 9 <split [1.4K/151]> Fold09 <tibble [2 × 3]> <tibble [0 × 1]>
## 10 <split [1.4K/151]> Fold10 <tibble [2 × 3]> <tibble [0 × 1]>
The results are similar to the folds results with some extra columns. The column .metrics contains the performance statistics created from the 10 assessment sets. These can be manually unnested but the tune package contains a number of simple functions that can extract these data:
collect_metrics(rf_fit_rs)
## # A tibble: 2 x 5
## .metric .estimator mean n std_err
## <chr> <chr> <dbl> <int> <dbl>
## 1 accuracy binary 0.833 10 0.0111
## 2 roc_auc binary 0.903 10 0.00842
Think about these values we now have for accuracy and AUC. These performance metrics are now more realistic (i.e. lower) than our ill-advised first attempt at computing performance metrics in the section above. If we wanted to try different model types for this data set, we could more confidently compare performance metrics computed using resampling to choose between models. Also, remember that at the end of our project, we return to our test set to estimate final model performance. We have looked at this once already before we started using resampling, but let’s remind ourselves of the results:
rf_testing_pred %>% # test set predictions
roc_auc(truth = class, .pred_PS)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 roc_auc binary 0.909
rf_testing_pred %>% # test set predictions
accuracy(truth = class, .pred_class)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.837
The performance metrics from the test set are much closer to the performance metrics computed using resampling than our first (“bad idea”) attempt. Resampling allows us to simulate how well our model will perform on new data, and the test set acts as the final, unbiased check for our model’s performance.
4. Tuning
Some model parameters cannot be learned directly from a data set during model training; these kinds of parameters are called hyperparameters. Some examples of hyperparameters include the number of predictors that are sampled at splits in a tree-based model (we call this mtry in tidymodels) or the learning rate in a boosted tree model (we call this learn_rate). Instead of learning these kinds of hyperparameters during model training, we can estimate the best values for these values by training many models on resampled data sets and exploring how well all these models perform. This process is called tuning.
We’ll need the following packages:
library(tidymodels) # for the tune package, along with the rest of tidymodels
# Helper packages
library(modeldata) # for the cells data
library(vip) # for variable importance plots
I. The cell image data, revisited
In the previous section (Evaluating), we introduced a data set of images of cells that were labeled by experts as well-segmented (WS) or poorly segmented (PS). We trained a random forest model to predict which images are segmented well vs. poorly, so that a biologist could filter out poorly segmented cell images in their analysis. We used resampling to estimate the performance of our model on this data.
II. Predicting image segmentation, but better
Random forest models are a tree-based ensemble method, and typically perform well with default hyperparameters. However, the accuracy of some other tree-based models, such as boosted tree models or decision tree models, can be sensitive to the values of hyperparameters. In this section, we will train a decision tree model. There are several hyperparameters for decision tree models that can be tuned for better performance. Let’s explore:
- the complexity parameter (which we call
cost_complexityin tidymodels) for the tree, and - the maximum
tree_depth.
Tuning these hyperparameters can improve model performance because decision tree models are prone to overfitting. This happens because single tree models tend to fit the training data too well — so well, in fact, that they over-learn patterns present in the training data that end up being detrimental when predicting new data.
We will tune the model hyperparameters to avoid overfitting. Tuning the value of cost_complexity helps by pruning back our tree. It adds a cost, or penalty, to error rates of more complex trees; a cost closer to zero decreases the number tree nodes pruned and is more likely to result in an overfit tree. However, a high cost increases the number of tree nodes pruned and can result in the opposite problem—an underfit tree. Tuning tree_depth, on the other hand, helps by stopping our tree from growing after it reaches a certain depth. We want to tune these hyperparameters to find what those two values should be for our model to do the best job predicting image segmentation.
Before we start the tuning process, we split our data into training and testing sets, just like when we trained the model with one default set of hyperparameters. As before, we can use strata = class if we want our training and testing sets to be created using stratified sampling so that both have the same proportion of both kinds of segmentation.
set.seed(123)
cell_split <- initial_split(cells %>% select(-case),
strata = class)
cell_train <- training(cell_split)
cell_test <- testing(cell_split)
We use the training data for tuning the model.
III. Tuning hyperparameters
Let’s start with the parsnip package, using a decision_tree() model with the rpart engine. To tune the decision tree hyperparameters cost_complexity and tree_depth, we create a model specification that identifies which hyperparameters we plan to tune.
tune_spec <-
decision_tree(
cost_complexity = tune(),
tree_depth = tune()
) %>%
set_engine("rpart") %>%
set_mode("classification")
tune_spec
## Decision Tree Model Specification (classification)
##
## Main Arguments:
## cost_complexity = tune()
## tree_depth = tune()
##
## Computational engine: rpart
Think of tune() here as a placeholder. After the tuning process, we will select a single numeric value for each of these hyperparameters. For now, we specify our parsnip model object and identify the hyperparameters we will tune().
We can’t train this specification on a single data set (such as the entire training set) and learn what the hyperparameter values should be, but we can train many models using resampled data and see which models turn out best. We can create a regular grid of values to try using some convenience functions for each hyperparameter:
tree_grid <- grid_regular(cost_complexity(),
tree_depth(),
levels = 5)
The function grid_regular() is from the dials package. It chooses sensible values to try for each hyperparameter; here, we asked for 5 of each. Since we have two to tune, grid_regular() returns 5 $\times$ 5 = 25 different possible tuning combinations to try in a tidy tibble format.
tree_grid
## # A tibble: 25 x 2
## cost_complexity tree_depth
## <dbl> <int>
## 1 0.0000000001 1
## 2 0.0000000178 1
## 3 0.00000316 1
## 4 0.000562 1
## 5 0.1 1
## 6 0.0000000001 4
## 7 0.0000000178 4
## 8 0.00000316 4
## 9 0.000562 4
## 10 0.1 4
## # … with 15 more rows
Here, you can see all 5 values of cost_complexity ranging up to r max(tree_grid$cost_complexity). These values get repeated for each of the 5 values of tree_depth:
tree_grid %>%
count(tree_depth)
## # A tibble: 5 x 2
## tree_depth n
## <int> <int>
## 1 1 5
## 2 4 5
## 3 8 5
## 4 11 5
## 5 15 5
Armed with our grid filled with 25 candidate decision tree models, let’s create cross-validation folds for tuning:
set.seed(234)
cell_folds <- vfold_cv(cell_train)
Tuning in tidymodels requires a resampled object created with the rsample package.
IV. Model tuning with a grid
We are ready to tune! Let’s use tune_grid() to fit models at all the different values we chose for each tuned hyperparameter. There are several options for building the object for tuning:
- Tune a model specification along with a recipe or model, or
- Tune a
workflow()that bundles together a model specification and a recipe or model preprocessor.
Here we use a workflow() with a straightforward formula; if this model required more involved data preprocessing, we could use add_recipe() instead of add_formula().
set.seed(345)
tree_wf <- workflow() %>%
add_model(tune_spec) %>%
add_formula(class ~ .)
tree_res <-
tree_wf %>%
tune_grid(
resamples = cell_folds,
grid = tree_grid
)
tree_res
## # Tuning results
## # 10-fold cross-validation
## # A tibble: 10 x 4
## splits id .metrics .notes
## <list> <chr> <list> <list>
## 1 <split [1.4K/152]> Fold01 <tibble [50 × 6]> <tibble [0 × 1]>
## 2 <split [1.4K/152]> Fold02 <tibble [50 × 6]> <tibble [0 × 1]>
## 3 <split [1.4K/152]> Fold03 <tibble [50 × 6]> <tibble [0 × 1]>
## 4 <split [1.4K/152]> Fold04 <tibble [50 × 6]> <tibble [0 × 1]>
## 5 <split [1.4K/152]> Fold05 <tibble [50 × 6]> <tibble [0 × 1]>
## 6 <split [1.4K/151]> Fold06 <tibble [50 × 6]> <tibble [0 × 1]>
## 7 <split [1.4K/151]> Fold07 <tibble [50 × 6]> <tibble [0 × 1]>
## 8 <split [1.4K/151]> Fold08 <tibble [50 × 6]> <tibble [0 × 1]>
## 9 <split [1.4K/151]> Fold09 <tibble [50 × 6]> <tibble [0 × 1]>
## 10 <split [1.4K/151]> Fold10 <tibble [50 × 6]> <tibble [0 × 1]>
Once we have our tuning results, we can both explore them through visualization and then select the best result. The function collect_metrics() gives us a tidy tibble with all the results. We had 25 candidate models and two metrics, accuracy and roc_auc, and we get a row for each .metric and model.
tree_res %>%
collect_metrics()
## # A tibble: 50 x 8
## cost_complexity tree_depth .metric .estimator mean n std_err .config
## <dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 0.0000000001 1 accuracy binary 0.734 10 0.00877 Model01
## 2 0.0000000001 1 roc_auc binary 0.772 10 0.00617 Model01
## 3 0.0000000178 1 accuracy binary 0.734 10 0.00877 Model02
## 4 0.0000000178 1 roc_auc binary 0.772 10 0.00617 Model02
## 5 0.00000316 1 accuracy binary 0.734 10 0.00877 Model03
## 6 0.00000316 1 roc_auc binary 0.772 10 0.00617 Model03
## 7 0.000562 1 accuracy binary 0.734 10 0.00877 Model04
## 8 0.000562 1 roc_auc binary 0.772 10 0.00617 Model04
## 9 0.1 1 accuracy binary 0.734 10 0.00877 Model05
## 10 0.1 1 roc_auc binary 0.772 10 0.00617 Model05
## # … with 40 more rows
We might get more out of plotting these results:
tree_res %>%
collect_metrics() %>%
mutate(tree_depth = factor(tree_depth)) %>%
ggplot(aes(cost_complexity, mean, color = tree_depth)) +
geom_line(size = 1.5, alpha = 0.6) +
geom_point(size = 2) +
facet_wrap(~ .metric, scales = "free", nrow = 2) +
scale_x_log10(labels = scales::label_number()) +
scale_color_viridis_d(option = "plasma", begin = .9, end = 0)
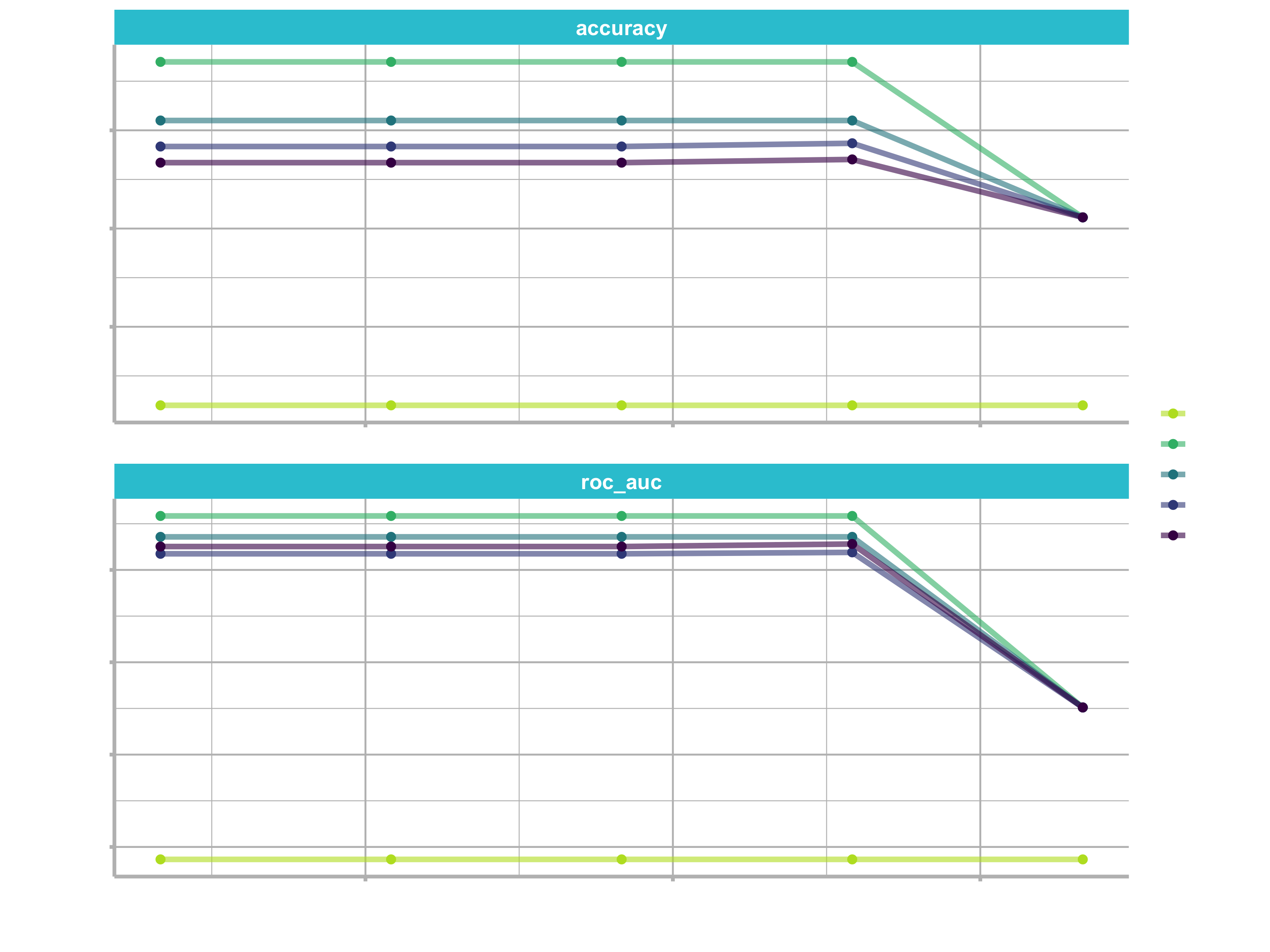
We can see that our “stubbiest” tree, with a depth of 1, is the worst model according to both metrics and across all candidate values of cost_complexity. Our deepest tree, with a depth of 15, did better. However, the best tree seems to be between these values with a tree depth of 4. The show_best() function shows us the top 5 candidate models by default:
tree_res %>%
show_best("roc_auc")
## # A tibble: 5 x 8
## cost_complexity tree_depth .metric .estimator mean n std_err .config
## <dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 0.0000000001 4 roc_auc binary 0.865 10 0.00965 Model06
## 2 0.0000000178 4 roc_auc binary 0.865 10 0.00965 Model07
## 3 0.00000316 4 roc_auc binary 0.865 10 0.00965 Model08
## 4 0.000562 4 roc_auc binary 0.865 10 0.00965 Model09
## 5 0.0000000001 8 roc_auc binary 0.859 10 0.0104 Model11
We can also use the select_best() function to pull out the single set of hyperparameter values for our best decision tree model:
best_tree <- tree_res %>%
select_best("roc_auc")
best_tree
## # A tibble: 1 x 3
## cost_complexity tree_depth .config
## <dbl> <int> <chr>
## 1 0.0000000001 4 Model06
These are the values for tree_depth and cost_complexity that maximize AUC in this data set of cell images.
V. inalizing our model+
We can update (or “finalize”) our workflow object tree_wf with the values from select_best().
final_wf <-
tree_wf %>%
finalize_workflow(best_tree)
final_wf
## ══ Workflow ══════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: decision_tree()
##
## ── Preprocessor ──────────────────────────────────────────────────────
## class ~ .
##
## ── Model ─────────────────────────────────────────────────────────────
## Decision Tree Model Specification (classification)
##
## Main Arguments:
## cost_complexity = 1e-10
## tree_depth = 4
##
## Computational engine: rpart
Our tuning is done!
VI. Exploring results
Let’s fit this final model to the training data. What does the decision tree look like?
final_tree <-
final_wf %>%
fit(data = cell_train)
final_tree
## ══ Workflow [trained] ════════════════════════════════════════════════
## Preprocessor: Formula
## Model: decision_tree()
##
## ── Preprocessor ──────────────────────────────────────────────────────
## class ~ .
##
## ── Model ─────────────────────────────────────────────────────────────
## n= 1515
##
## node), split, n, loss, yval, (yprob)
## * denotes terminal node
##
## 1) root 1515 540 PS (0.64356436 0.35643564)
## 2) total_inten_ch_2< 47256.5 731 63 PS (0.91381669 0.08618331)
## 4) total_inten_ch_2< 37166 585 19 PS (0.96752137 0.03247863) *
## 5) total_inten_ch_2>=37166 146 44 PS (0.69863014 0.30136986)
## 10) avg_inten_ch_1< 99.15056 98 14 PS (0.85714286 0.14285714) *
## 11) avg_inten_ch_1>=99.15056 48 18 WS (0.37500000 0.62500000)
## 22) fiber_align_2_ch_3>=1.47949 20 8 PS (0.60000000 0.40000000) *
## 23) fiber_align_2_ch_3< 1.47949 28 6 WS (0.21428571 0.78571429) *
## 3) total_inten_ch_2>=47256.5 784 307 WS (0.39158163 0.60841837)
## 6) fiber_width_ch_1< 11.19756 329 137 PS (0.58358663 0.41641337)
## 12) avg_inten_ch_1< 194.4183 254 82 PS (0.67716535 0.32283465) *
## 13) avg_inten_ch_1>=194.4183 75 20 WS (0.26666667 0.73333333)
## 26) total_inten_ch_3>=62458.5 23 9 PS (0.60869565 0.39130435) *
## 27) total_inten_ch_3< 62458.5 52 6 WS (0.11538462 0.88461538) *
## 7) fiber_width_ch_1>=11.19756 455 115 WS (0.25274725 0.74725275)
## 14) shape_p_2_a_ch_1>=1.225676 300 97 WS (0.32333333 0.67666667)
## 28) avg_inten_ch_2>=362.0108 55 23 PS (0.58181818 0.41818182) *
## 29) avg_inten_ch_2< 362.0108 245 65 WS (0.26530612 0.73469388) *
## 15) shape_p_2_a_ch_1< 1.225676 155 18 WS (0.11612903 0.88387097) *
This final_tree object has the finalized, fitted model object inside. You may want to extract the model object from the workflow. To do this, you can use the helper function pull_workflow_fit().
For example, perhaps we would also like to understand what variables are important in this final model. We can use the vip package to estimate variable importance.
library(vip)
final_tree %>%
pull_workflow_fit() %>%
vip()
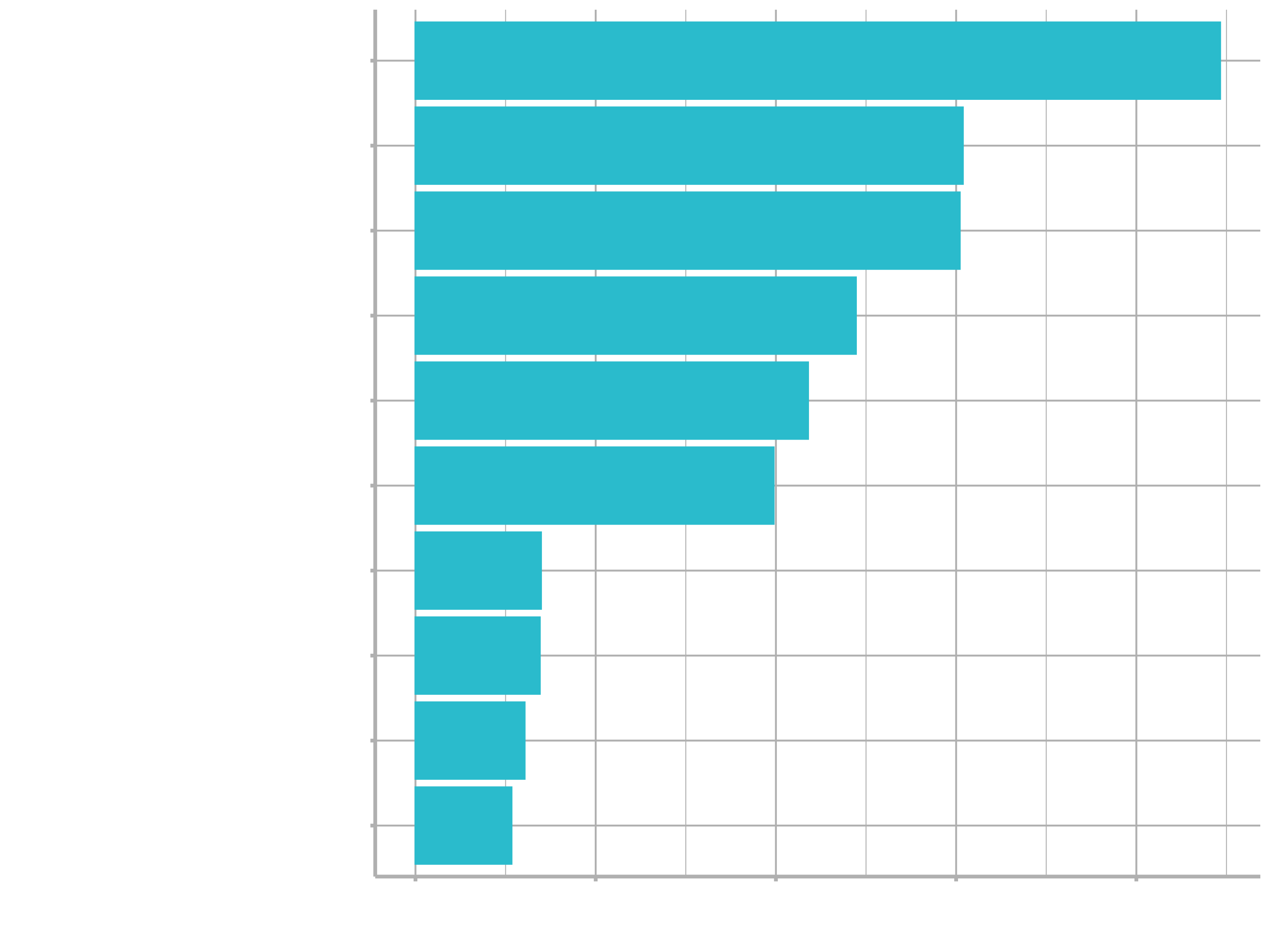
These are the automated image analysis measurements that are the most important in driving segmentation quality predictions.
VII. The last fit
Finally, let’s return to our test data and estimate the model performance we expect to see with new data. We can use the function last_fit() with our finalized model; this function fits the finalized model on the full training data set and evaluates the finalized model on the testing data.
final_fit <-
final_wf %>%
last_fit(cell_split)
final_fit %>%
collect_metrics()
## # A tibble: 2 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.802
## 2 roc_auc binary 0.860
final_fit %>%
collect_predictions() %>%
roc_curve(class, .pred_PS) %>%
autoplot()
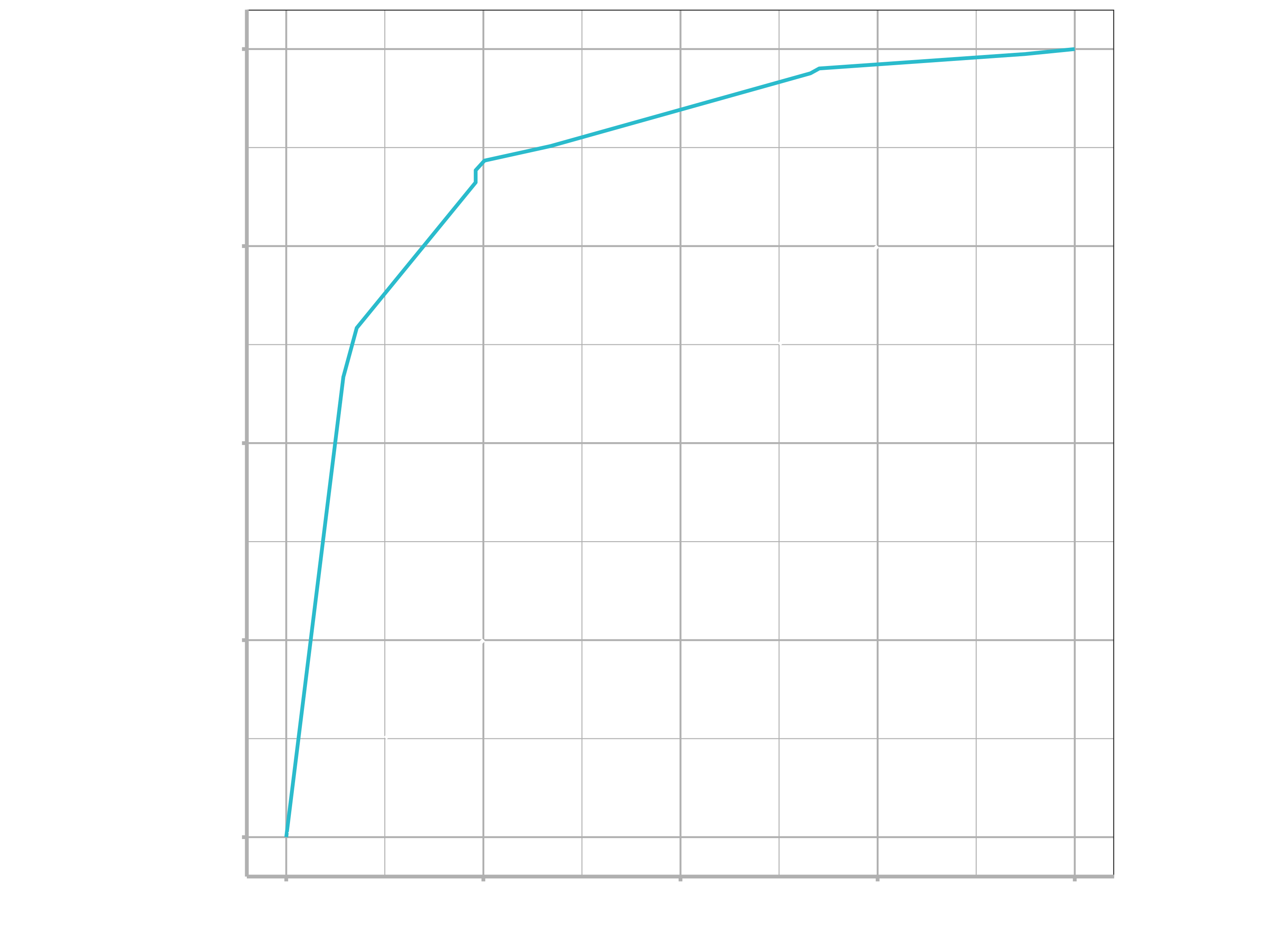
The performance metrics from the test set indicate that we did not overfit during our tuning procedure.
I leave it to you to explore whether you can tune a different decision tree hyperparameter. You can use the args() function to see which parsnip object arguments are available:
args(decision_tree)
## function (mode = "unknown", cost_complexity = NULL, tree_depth = NULL,
## min_n = NULL)
## NULL
You could tune the other hyperparameter we didn’t use here, min_n, which sets the minimum n to split at any node. This is another early stopping method for decision trees that can help prevent overfitting. Use this searchable table to find the original argument for min_n in the rpart package (hint). See whether you can tune a different combination of hyperparameters and/or values to improve a tree’s ability to predict cell segmentation quality.
Business case
Because this session contains already a lot of new information to process, there will be no business case and challenge this time. Hence, this class is split into two parts. Next week, I will explain a business case (with the canyon bike date) and you can get your hands on further machine learning. The following problem will be part of the next session:
Problem definition
- Which Bike Categories are in high demand?
- Which Bike Categories are under represented?
- GOAL: Use a pricing algorithm to determine a new product price in a category gap
Challenge
See above.
In practice, some algorithms (i.e. R’s
glmnet::glmnet()implement standardization internally and re-scale prior to returning term estimates and predictions. This means that features need not be scaled prior to use. ↩︎Feature Importance can be obtained with additional methods for global (variable importance) and local (e.g. LIME) model understanding. ↩︎
In practice, some algorithms (i.e. R’s
kernlabs::ksvm()implement standardization internally and re-scale prior to returning term estimates and predictions. This means that features need not be scaled prior to use. ↩︎







